Inversion: Temporal Pincer in Cancer Genome
Head-to-head (5' to 5') and tail-to-tail (3' to 3') inversion breakpoints explained.
1. Opening
I’m a big fan of the movie Tenet, especially the mind-bending concept of temporal pincers. Without giving too much away, let’s just say the plot goes forwards and backwards - kind of like a cancer genome when a somatic inversion rewrites the script.
In the world of bioinformatics, I occasionally come across a temporal pincer in a cancer genome - an inverted DNA fragment. Recently, I rehashed my understanding of head-to-head (5’ to 5’) and tail-to-tail (3’ to 3’) inversion breakpoints. Here, I share my notes on what they are and how to spot them in sequencing alignments. Below are some online resources I’ve checked out to learn about inversions:
2. What is Inversion?
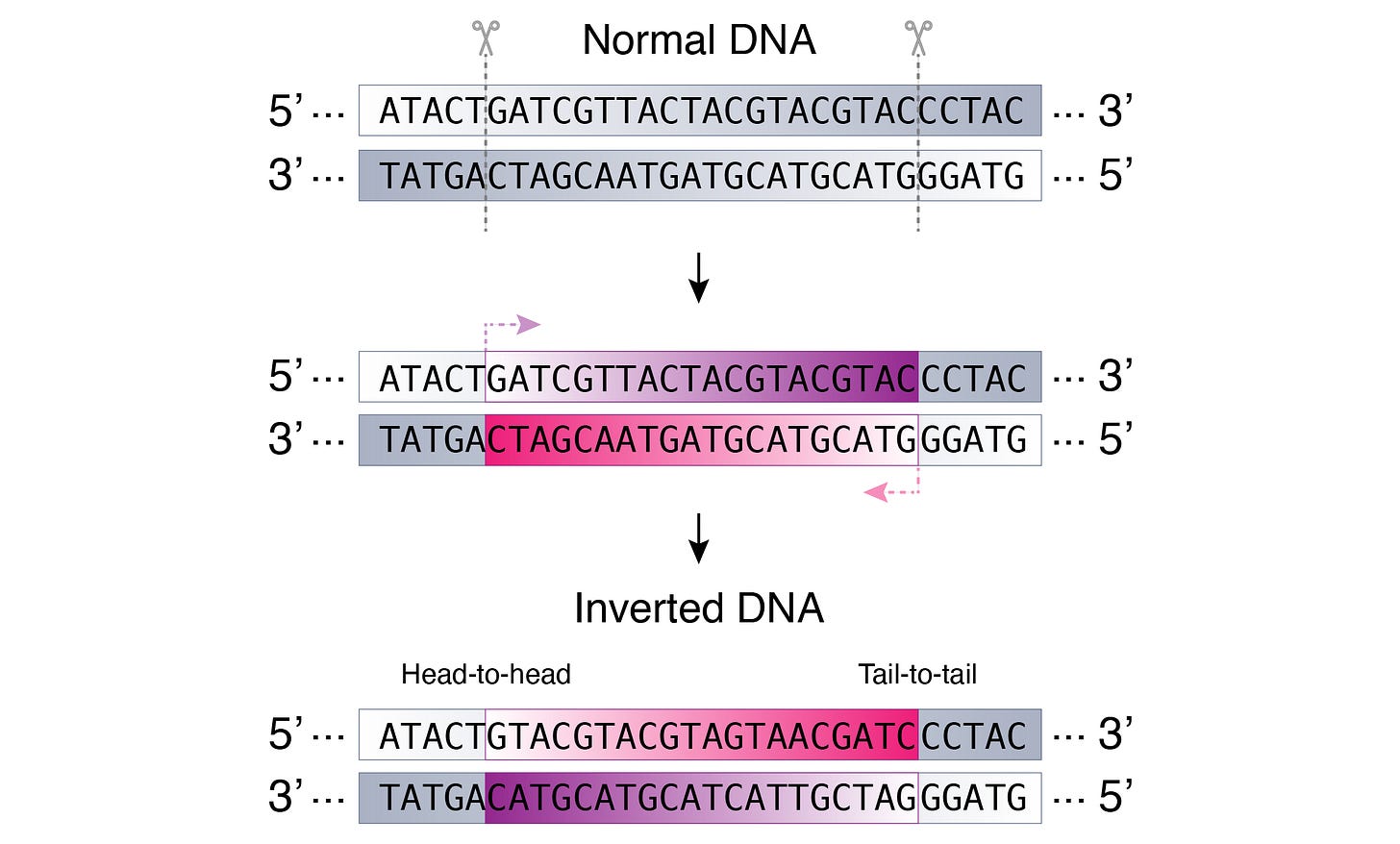
An inversion is a structural DNA variant whereby a DNA fragment is flipped in the opposite direction within the genome (Figure 1).
Notably, the inverted DNA is inserted back into the genome in the reverse-complement direction. This is to preserve the 5’ and 3’ orientation in the DNA double helix structure, as shown above.
Inversions are important to understand because, in cancer, they can be early structural alterations that initiate tumorigenesis.
3. Head-to-Head and Tail-to-Tail Breakpoints
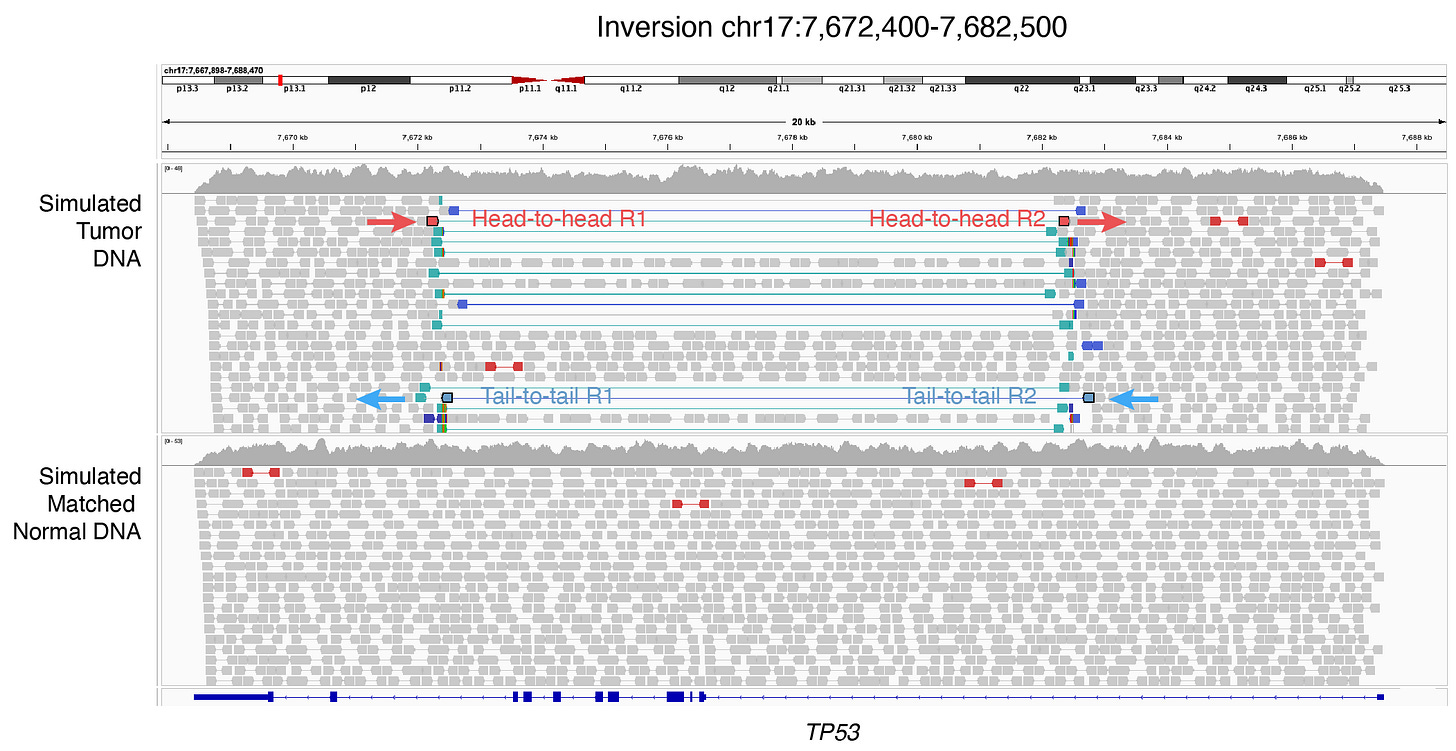
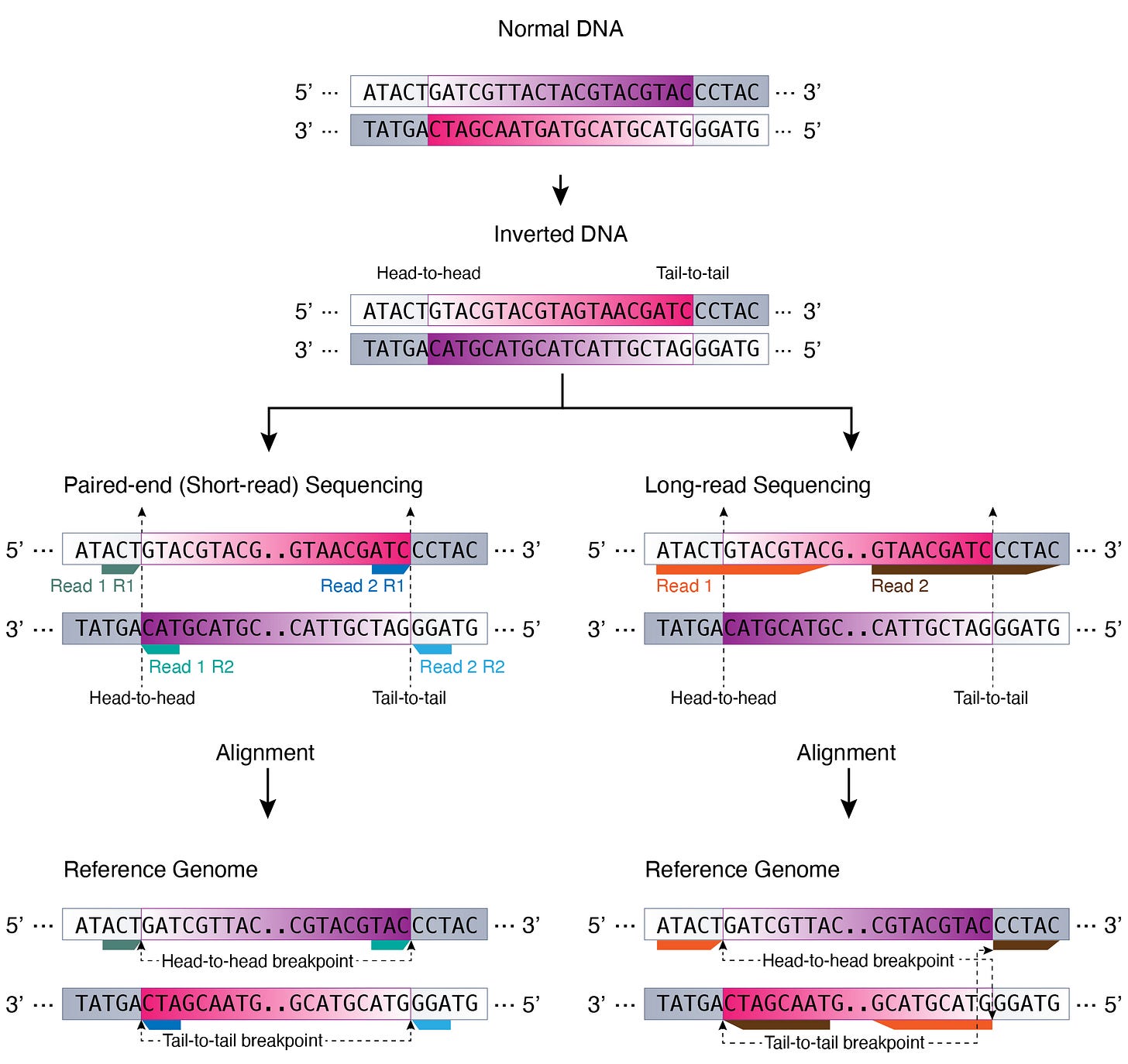
An inversion can be long, long enough that sequencing reads cannot capture the whole event end-to-end. But, what’s happened, happened. Sequencing reads should still be able to observe parts of the inversion. This is where the head-to-head and tail-to-tail breakpoints come in (Figure 2). As shown below, a read that captures the 5’ ends (5’ to 5’) of the inversion generates a head-to-head breakpoint upon alignment to the reference genome while a read that captures the 3’ ends (3’ to 3’) leads to a tail-to-tail breakpoint.
4. Inversion by Sequencing Technologies
Let’s take a look at the inversion alignment patterns between short-read (paired-end) and long-read sequencing data.
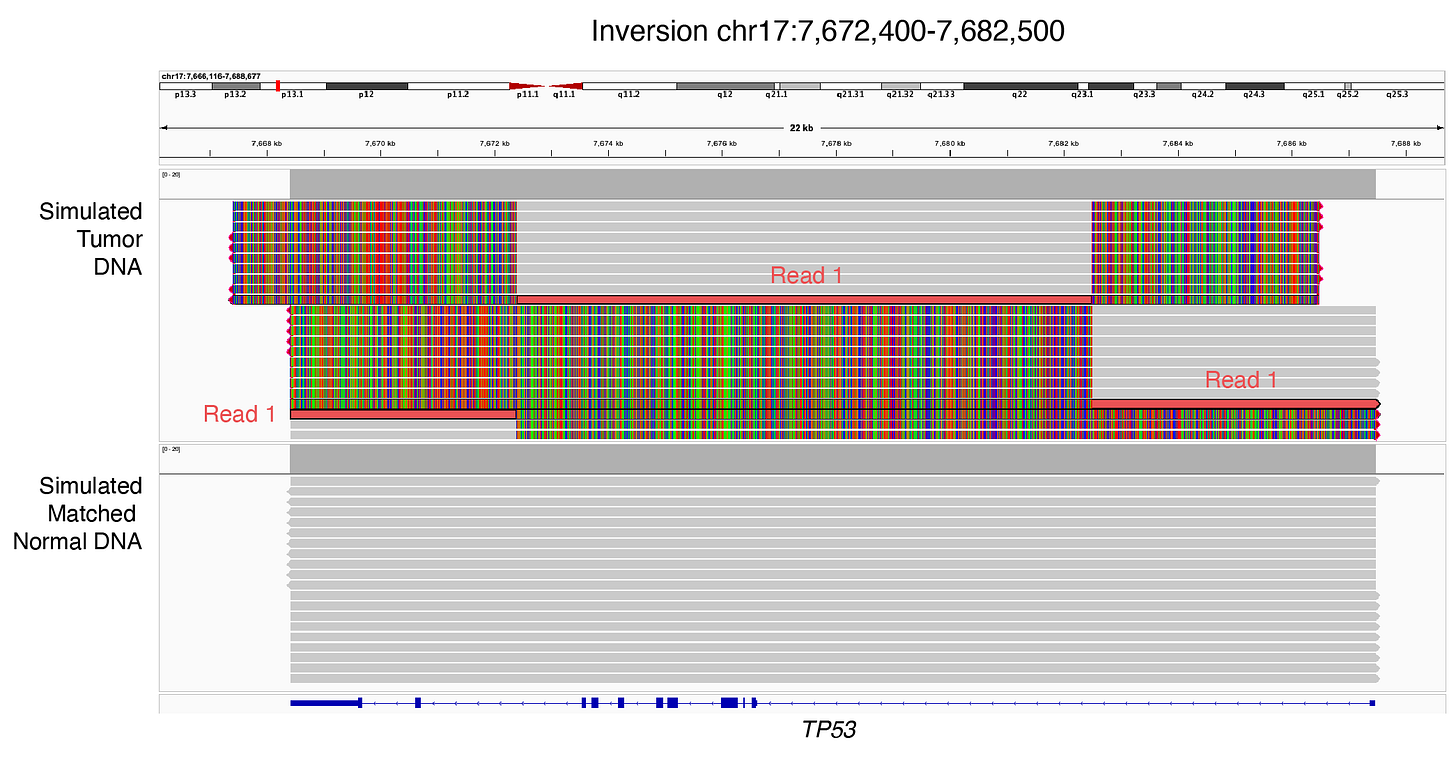
Here, I simulated a somatic inversion (chr17:7,672,400-7,682,500) in the hg38 genomic region that encodes the tumor-suppressor gene TP53, and generated both short-read (151-bp) and long-read (~10,000-bp) DNA sequencing data for the same event.
Paired-end (Short-read) Sequencing
Owing to the short read length, short-read paired-end sequencing cannot capture most inversion events end-to-end. As we saw above, this results in observing 2 kinds of read alignments: head-to-head (5’ to 5’) alignment (red) and tail-to-tail (3’ to 3’) alignment (blue) (Figure 3).
Long-read Sequencing
If a long read captures an inversion event end-to-end, the read will have 2 supplementary alignments (soft-clipped) and have 3 alignment records (Figure 4).
5. Inversion Representation
In my previous post, I introduced the Occam’s Variant Grammar. If we identified variants in both the short-read and long-read data, we would have found the following variants encoded in the Grammar:
// 5' to 5' (head-to-head) variant
dna_variant_1 = {
chromosome_1: 'chr17',
position_1: 7672399,
strand_1: '+',
operation_1: 'D', // downstream (towards 3')
chromosome_2: 'chr17',
position_2: 7682500,
strand_2: '-',
operation_2: 'D', // downstream (towards 3')
sequence: ''
}
// 3' to 3' (tail-to-tail) variant
dna_variant_2 = {
chromosome_1: 'chr17',
position_1: 7672400,
strand_1: '-',
operation_1: 'U', // upstream (towards 5')
chromosome_2: 'chr17',
position_2: 7682501,
strand_2: '+',
operation_2: 'U', // upstream (towards 5')
sequence: ''
} To detect the inversion, a variant caller that uses the Occam’s Variant Grammar would look for the above pair of events and classify them as evidence supporting an inversion.
Stay tuned for more on how structural variants are implicated at the RNA and peptide levels.
6. Closing
Haven’t you guessed by now? This whole blog post is a temporal pincer. See if you can spot famous lines from Tenet in this post and let us know in the comments section below! I gave you one to begin with.
“I’ll see you at the beginning, friend.”
7. Acknowledgement
This work was made possible through the generous support of the Jaime Leandro Foundation, a non-profit foundation committed to brining therapeutic cancer vaccines to patients.