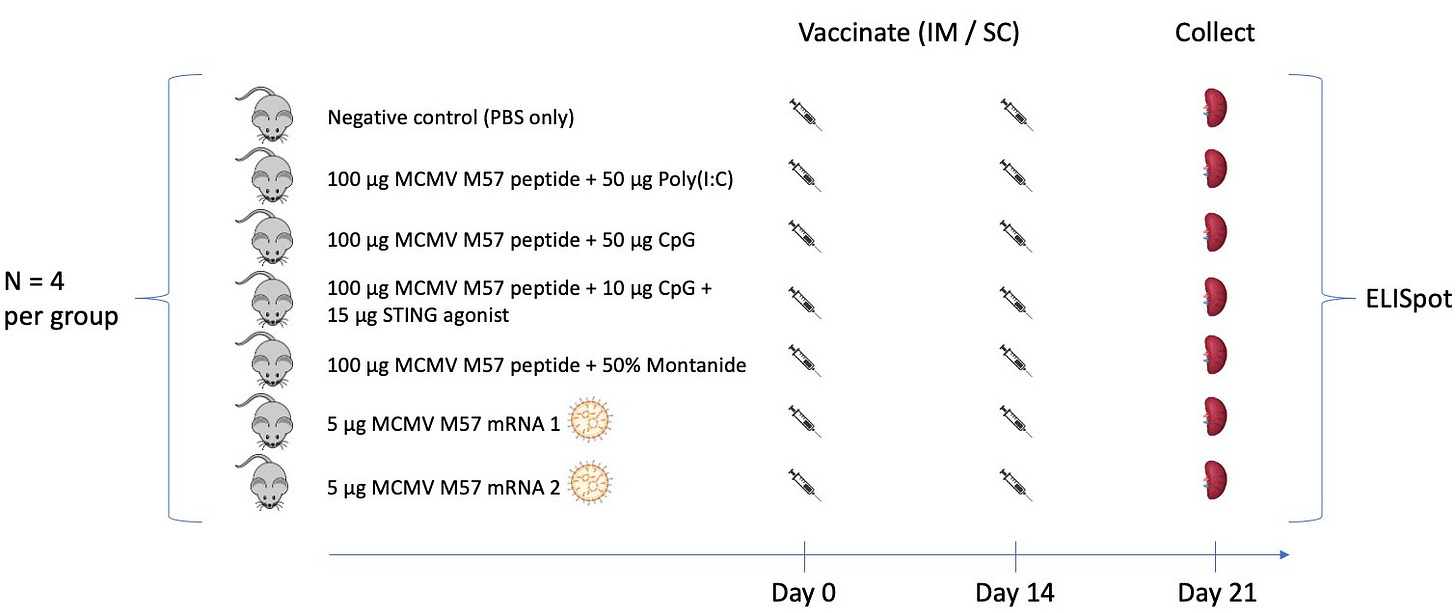
An experiment comparing the potency of vaccine formulations in C57BL/6 mice
Introducing the Optimizing Immunization Strategies for Therapeutic Efficacy and Response (OISTER) project
One challenge in optimizing cancer vaccines is our incomplete understanding of comparative vaccine potency. To address this, our lab is on a somewhat quixotic journey to benchmark cancer vaccine formulations and dosing schedules using functional readouts of T cell and B cell responses. Amongst a sea of competing formulations in clinical trials and preclinical development (many variations of peptide + adjuvant vaccines, DNA vaccines, mRNA vaccines, dendritic cell vaccines, viral component vaccines, etc.), we want to know which formulations are most immunogenic and why. In an early stab at this, we set up an experiment to vaccinate C57BL/6 mice with a known immunogenic MCMV epitope (M57) formulated as peptide + adjuvant and mRNA vaccines as below:
Here we tested poly(I:C), CpG, and montanide (all relatively common vaccine adjuvants), a STING agonist (which we can’t specify for now given constraints in our MTA with the supplier), and two mRNA vaccine formulations. For each of the mRNA formulations, N1-Methylpseudouridine was substituted for uridine. mRNA 2 included a segment coding for an MHC class I signal peptide as well as the vaccine epitope, whereas mRNA 1 did not. mRNA was carried by lipid nanoparticles that were manufactured in our lab using the Moderna mRNA-1273 lipid mix. All vaccine formulations were given intramuscularly, except the STING agonist which was given subcutaneously. The vaccine schedule was a simple prime and boost design, with a single priming dose given on day 0, a boost dose given on day 14, and splenocytes analyzed for M57-specific T cell responses by interferon gamma ELISpot on day 21. Experiments were done by Maria Sambade and Jess Alley, who wrote about generating the mRNA/LNP vaccines here. Results:

CpG performed best out of the adjuvants tested for peptide vaccines, however mRNA/LNP vaccines may have performed best overall. We will need to repeat this experiment with more animals per group and test other functional readouts along with interferon gamma secretion to be sure. Notably, poly(I:C) and montanide both performed rather poorly here. Most human clinical trials that use poly(I:C) actually use poly(I:C)-LC (Hiltonol), a poly(I:C) variant that is augmented by addition of polylysine. We need to test poly(I:C)-LC in future comparisons for translational relevance. We tested the STING agonist only with CpG here because (1) we had a limited amount of it, and (2) we had seen in prior experiments that CpG was a better adjuvant than poly(I:C) in this system.
Going forward, we plan to test other vaccine formulations (more adjuvants and DC vaccines next, hopefully DNA vaccines after that), different vaccine schedules (varying prime and boost timing and number of injections), and a set of validated neoantigens (rather than the M57 antigen alone) in tumor-bearing mice. We will continue to post results of these experiments here, and we hope this will be useful to the cancer vaccine community as we all try to figure out what are the best formulations to take to clinical trials.
A critical variable not addressed here is how to best do tumor antigen selection. Vaccine immunogenicity is irrelevant if the vaccine antigens are not presented by the tumor cells (just like antigen selection is irrelevant if vaccine immunogenicity is poor). More to come on that in future posts…
Many thanks to Natera for providing funding for this work along with Jamie Leandro Foundation (JLF), a non-profit foundation dedicated to making therapeutic cancer vaccines available for patients who could benefit from them. Thanks to both Natera and JLF for supporting publication of these results on Substack rather waiting for a “complete story” to emerge and be published as a preprint or peer-reviewed article.